General Description
Inorganic Pyrophosphatase (PPase) is a highly active enzyme from a recombinant source in E. coli provided as a liquid reagent, manufactured under GMP process requirements and tested extensively for the absence of nucleases, alkaline phosphatase and other contaminants. PPase is an important biological catalyst because it has the unique ability to catalyze the hydrolysis of a high energy molecule. It is widely used as an auxiliary enzyme to improve the efficiency of many reactions in molecular biology and biotechnology.
Mechanism of Action
PPase catalyzes a thermodynamically favorable reaction, the hydrolysis of inorganic pyrophosphate (PPi) to two molecules of orthophosphate (Pi) ions. Since this reaction is rapid, PPi, which is a common byproduct and potent inhibitor of many polymerization reactions (including many involving RNA polymerases, DNA polymerases and reverse transcriptases) is continuously removed, driving the otherwise thermodynamically unfavorable transformation to completion and increasing the total yield of the primary reaction.
Application
PPase is used in applications where nucleic acid synthesis or amplification is prone to accumulation of pyrophosphate. The most common applications are in vitro transcription (IVT) processes and reverse transcription reactions, which show a significant increase in RNA yield when PPase is used. PPase is also used routinely to improve the efficiency of DNA replication, PCR, single-base extension reactions, DNA sequencing, multiply-prime rolling circle amplification and some specialized enzymatic assays.
Soluble Family I, Family II and membrane-integral (M-)Soluble inorganic pyrophosphatases (PPases) all hydrolyse pyrophosphate. Family I enzymes are ubiquitous hexameric or dimeric, utilise two Mg2+ to activate associative water attack and are potently inhibited by fluoride. Family II enzymes are found in bacteria and archaea and are DHH-superfamily dimers, utilise three metals and have a dissociative, metaphosphate-like transition state. Both soluble classes merely use PPi removal to drive biosynthesis. M-PPases are 16-helix membrane homodimers that couple PPi hydrolysis to Na+ and/or H+ pumping. Pumping is reversible and essential in plants, protozoa and many bacteria under energy stress.
Catalytically, M-PPases are 100-fold slower, bind fluoride weakly and seem to activate water only after PPi binding closes the active-site loop and displaces TM11-12 downward. This movement opens the ion gate, which suggests a binding-change model in which the pump stroke precedes hydrolysis, unifies Na+ and H+ transport and obviates the need for the hydrolytic proton to cross the funnel. Thus, while all PPases split the same substrate, they have evolved unrelated structures and distinct chemical strategies and only M-PPases convert PPi bond energy to membrane potential.
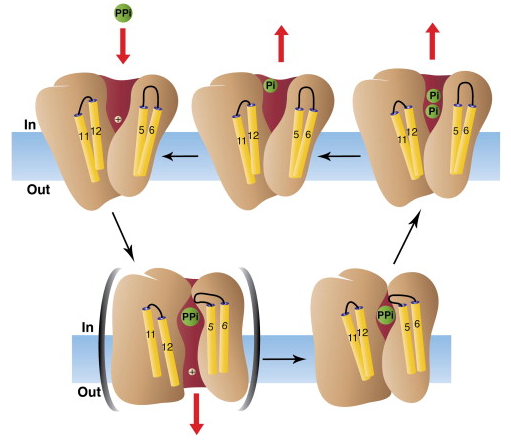 Fig. 1 The catalytic cycle of Inorganic Pyrophosphatase. (Kajander T.; et al. 2013)
Fig. 1 The catalytic cycle of Inorganic Pyrophosphatase. (Kajander T.; et al. 2013)
References
- Kajander T, et al. Inorganic pyrophosphatases: one substrate, three mechanisms. FEBS letters, 2013, 587(13): 1863-1869.
What is the source of Inorganic Pyrophosphatase?
Recombinant expression in E. coli.
Is your Inorganic Pyrophosphatase produced under GMP conditions?
Yes, it is manufactured following GMP process requirements.
Can Inorganic Pyrophosphatase be used in PCR?
Suitable for PCR and single-base extension reactions.
Can Inorganic Pyrophosphatase be used in DNA sequencing?
Yes, frequently used in DNA sequencing reactions.

 Fig. 1 The catalytic cycle of Inorganic Pyrophosphatase. (Kajander T.; et al. 2013)
Fig. 1 The catalytic cycle of Inorganic Pyrophosphatase. (Kajander T.; et al. 2013)













